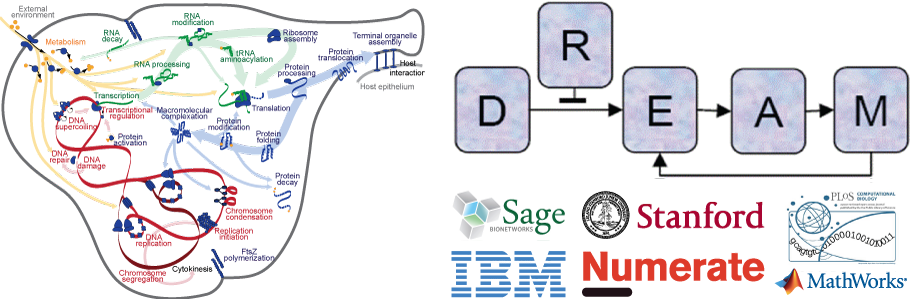
A central challenge in biology is to understand how phenotype arises from genotype. Despite decades of research which have produced vast amounts of biological data, a complete, predictive understanding of biological behavior remains elusive. Computational techniques are needed to assemble the rapidly growing amount of biological data into a unified understanding. Recently, researchers at Stanford University developed the first comprehensive dynamical “whole-cell” model of a living organism (Karr et al., 2012). The model broadly predicts the cell cycle dynamics of the Gram-positive bacterium M. genitalium from the level of individual molecules and their interactions, including its metabolism, transcription, translation, and replication. The model is composed of 28 sub-models of distinct cellular processes which were independently modeled at short time scales, and integrated together at longer time scales. The model was validated by broadly comparing its predictions to a wide range of experimental data across several biological processes and scales.
The goal of this Challenge is to explore and compare innovative approaches to parameter estimation of large, heterogeneous computational models. Participants are encouraged to develop and/or apply optimization methods, including the selection of the most informative experiments. The organizers encourage participants to form teams to collaboratively solve the Challenge.
Related publication:
Karr JR, Williams AH, Zucker JD, Raue A, Steiert B, Timmer J, et al. (2015) Summary of the DREAM8 Parameter Estimation Challenge: Toward Parameter Identification for Whole-Cell Models. PLoS Comput Biol 11(5): e1004096. https://doi.org/10.1371/journal.pcbi.1004096